Open-ST is published!
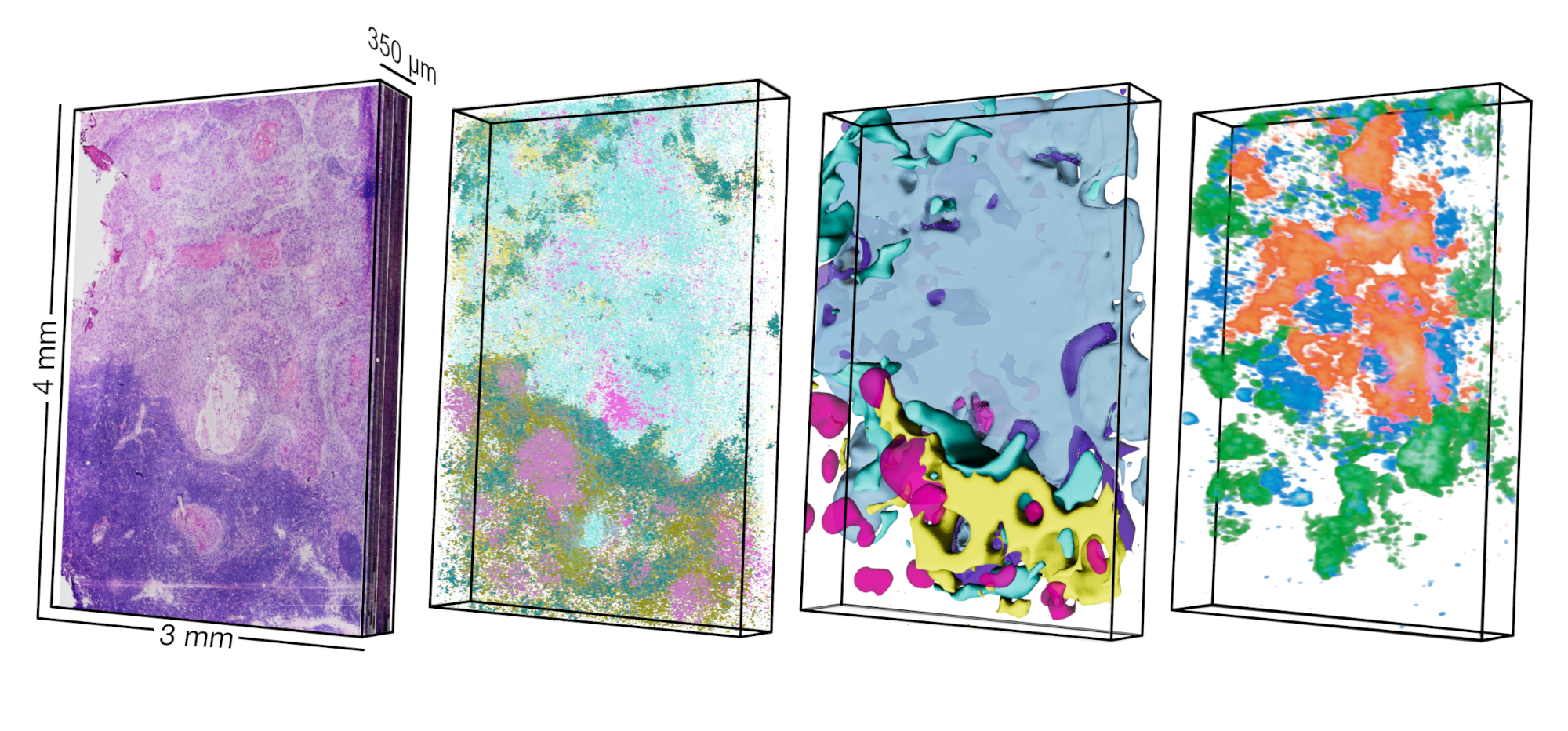
Our paper on presenting a new spatial transcriptomics platform is now published!
Published on June 24, 2024 by N. Rajewsky Lab
publication spatial transcriptomics openst methods development virtual tissue block lymph node cancer
0 min READ
We developed an inexpensive, do-it-yourself, high-resolution, and open source method to quantify gene expression directly in tissues (Marie Schott, Daniel León Perinán, Elena Splendiani et al. Open-ST: High-resolution spatial transcriptomics in 3D, Cell, 2024). We constructed the first high-resolution molecular tumor atlas in 3D (from a single patient) and learned from these ~ 1 million sequenced cells (in tissue space) which pathways and gene programs drive phenotypes of the primary and metastatic tumor. We are currently in the lab routinely generating spatial transcriptomic data as a highly informative readout for how cells communicate within tissues/tumors and how they react to perturbations (disease mutations, perturbations from the environment (viral infection, microplastics, …).